Database query results in extraction of corresponding information from input; it is vital to information-rich diciplines like genomics and molecular biology. As a software project in molecular biology, bioconductor includes a large portion of offline databases that are “bundled” as annotation packages. These annotation packages are standardised uisng a grammar defined by AnnotationDbi.
The grammar consists a main function AnnotationDbi::select and the four arguments it contains are
- x: the database names (as AnnotationDb object).
- keys: input.
- keytype: input types.
- colunms: outype type.
To a beginner of databases (such as Ensembl or NCBI), some difficulties in using these annotation packages.
- identify the “keytype” of an given input, espcially in the case of ID/accession number, is not trivial; database literacy needs to be there.
- when the information we need is not within a same annotation packages/databases, multiple databases are required and key variables that common among packages are required.
- During the learning process, I feel the R annotation package authors assume the reader to be expert in the databases while the databases manual touches little about API. So the related information are not usually systematic.
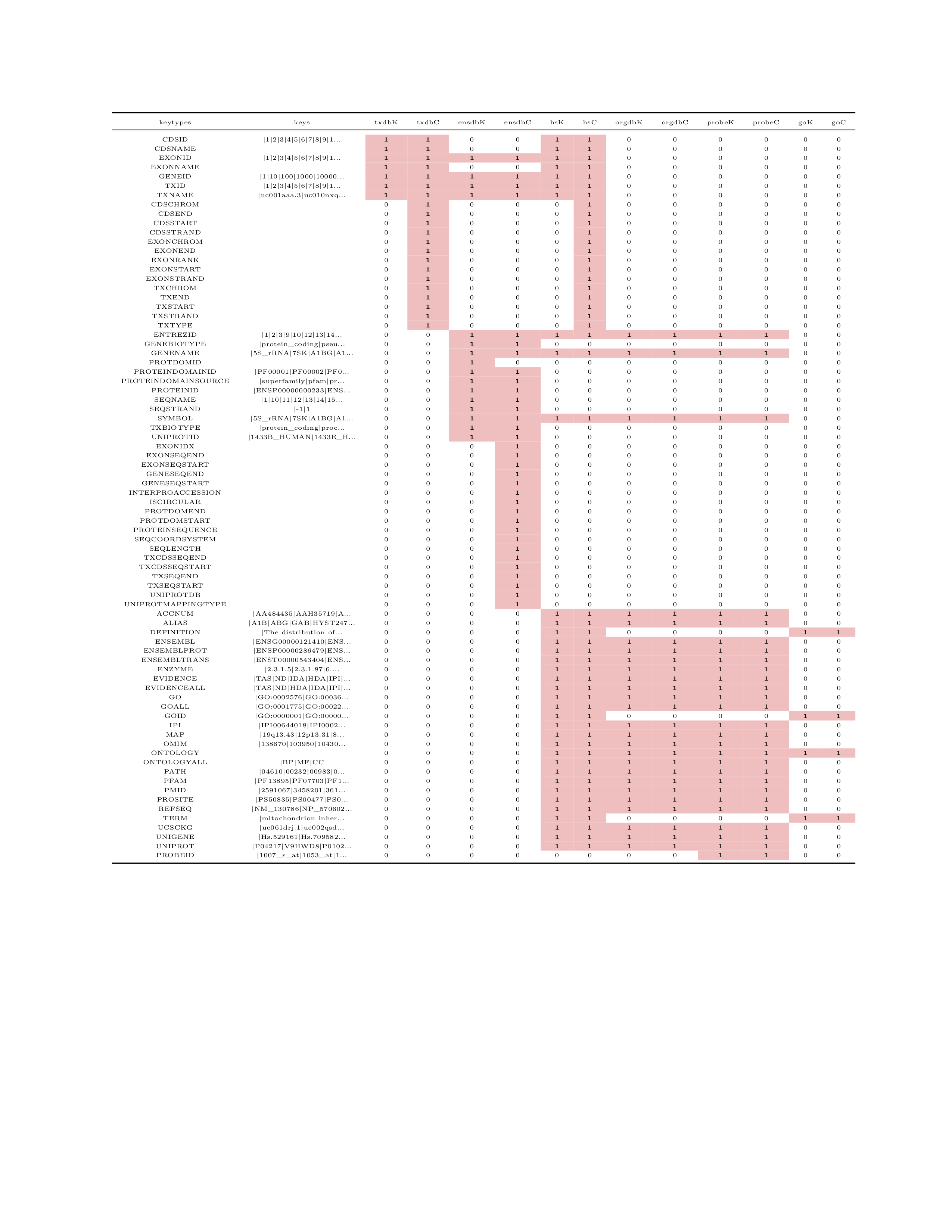
This blog summarises the information of the four arguments for annotation packages that are based on human hg19/GRCh37 genome construct. The reason why choosing the 2009 version instead of 2013 version (hg38/GRCh38) is because the summary was initially served as a cheat sheet for a project related to Affy U133 plus2 probeset and this Affy is designed based on the old human genome construct.
x <- c("tidyverse", "TxDb.Hsapiens.UCSC.hg19.knownGene", "EnsDb.Hsapiens.v75", "Homo.sapiens", "org.Hs.eg.db", "hgu133plus2.db", "GO.db", "kableExtra") ## AnnotationHub
y <- lapply(x, function(n){suppressMessages(library(n, character.only = TRUE))})
# extracting the keytypes (input type) and colums (output types) of all databases
txdb_k = keytypes(TxDb.Hsapiens.UCSC.hg19.knownGene)
txdb_c = columns(TxDb.Hsapiens.UCSC.hg19.knownGene)
ensdb_k = keytypes(EnsDb.Hsapiens.v75)
ensdb_c = columns(EnsDb.Hsapiens.v75)
hs_k = keytypes(Homo.sapiens)
hs_c = columns(Homo.sapiens)
orgdb_k = keytypes(org.Hs.eg.db)
orgdb_c = columns(org.Hs.eg.db)
probe_k = keytypes(hgu133plus2.db)
probe_c = columns(hgu133plus2.db)
go_k = keytypes(GO.db)
go_c = columns(GO.db)
iden <- list(txdb_k, txdb_c,
ensdb_k, ensdb_c,
hs_k, hs_c,
orgdb_k, orgdb_c,
probe_k, probe_c,
go_k, go_c)
## obtain unique keys/columns
rn <- unique(unlist(iden))
cn <- c("txdbK", "txdbC",
"ensdbK", "ensdbC",
"hsK", "hsC",
"orgdbK", "orgdbC",
"probeK", "probeC",
"goK", "goC")
## a null matrix
mat <- matrix(nrow = length(rn), ncol = length(cn),dimnames = list(rn, cn))
## fill the keytype/column to the corresponding database. assigning 1 for presence.
iden_length <- lapply(iden, function(n){
length(n)
})
iden_c <- c()
for(i in seq_along(cn)){
iden_c <- c(iden_c, rep.int(cn[i], times = iden_length[[i]]))
}
df <- tibble(from = iden_c, identifiers = unlist(iden))
for (i in seq_along(1:nrow(df))){
mat[which(rownames(mat) == df[[i,2]]),
which(colnames(mat) == df[[i,1]])] <- 1
}
mat[which(is.na(mat),arr.ind = TRUE)] <- 0A column called keys is constructed to list down the first few elements of each keytypes.
## concatenate the first a few element of the keys from each keytypes together seperated by \vert.
kc <- vector(mode = "character", length = 0L)
conc_descri <- function(key_string, max_length = 20){
## concatinate the first a few keys as the description of key types
concat_string <- ""
for (ks in seq_along(1:length(key_string))){
ori_string <- concat_string
if (!is.na(key_string[ks])){
concat_string <- stringi::stri_paste(concat_string, key_string[ks], sep = "|")
}
if (nchar(concat_string) >= max_length) {
## concat_string <- ori_string
concat_string <- strtrim(concat_string, max_length) %>%
paste0(., "...")
break
}
}
return(concat_string)
}
for(i in 1:nrow(mat)){
row_df <- mat[i, , drop = FALSE] == 1
row_df_colnames <- colnames(row_df)[min(which(row_df)[which(row_df)%%2 == 1])]
if (!is.na(row_df_colnames) && rownames(row_df) != "ONTOLOGY" && rownames(row_df) != "PROTDOMID"){ ## these two keytypes has problems
if(row_df_colnames == "txdbK"){
k <- keys(TxDb.Hsapiens.UCSC.hg19.knownGene, keytype = rownames(row_df))
} else if (row_df_colnames == "ensdbK"){
k <- keys(EnsDb.Hsapiens.v75, keytype = rownames(row_df))
} else if (row_df_colnames == "hsK"){
k <- keys(Homo.sapiens, keytype = rownames(row_df))
} else if (row_df_colnames == "orgdbK"){
k <- keys(org.Hs.eg.db, keytype = rownames(row_df))
} else if (row_df_colnames == "probeK"){
k <- keys(hgu133plus2.db, keytype = rownames(row_df))
} else if (row_df_colnames == "goK"){
k <- keys(GO.db, keytype = rownames(row_df))
} else {
k <- ""
}
kc[i] <- conc_descri(key_string = k)
} else {
kc[i] <- ""
}
} The cheat sheet was knitted to PDF with the following code and you can  download the cheatsheet.
download the cheatsheet.
################ latex code added before this R block ##############
# \newpage
# \pagenumbering{gobble}
################ end ###############################################
mat %>%
as_tibble(rownames = "keytypes")%>%
mutate_if(is.numeric, function(n){
ifelse(n == 1,
cell_spec(n, "latex", bold = T, background = "pink"),
n)
}) %>%
bind_cols(., tibble(keys = Hmisc::latexTranslate(kc))) %>% # add escape for latex output.
dplyr::select(keytypes, keys, everything()) %>%
kable(., format = "latex", escape = F, booktabs = T, linesep = "", align = "c") %>%
kable_styling(font_size = 4, latex_options = c("hold_position"))